
Introduction
The Bird 10,000 Genomes (B10K) Project is an initiative to generate representative draft genome sequences from all extant bird species. The establishment of this project is built on the success of the previous ordinal level project , which provided the first proof of concept for carrying out large-scale sequencing of multiple representative species across a vertebrate class and a window into the types of discoveries that can be made with such genomes. The announcement of the B10K Project was published on 3rd June 2015 in Nature .
The extraordinary diversity of Galapagos finches discovered during the 1826 voyage of the H.M.S. Beagle had an enormous impact on Darwin’s thinking about ‘On the Origin of Species’. His later studies on domesticated pigeons further inspired the development of the theory of evolution. Since then, bird studies have led to numerous pioneering findings that have established many new disciplines in biology, including Wallace’s biogeography, Mayr’s synthesis of speciation, MacArthur and Wilson’s island biogeography, Tinbergen’s ethology, and Hamilton’s kinship theory. With about 10,500 living species, birds constitute the most speciose class of terrestrial vertebrates and are adapted to and distributed throughout most terrestrial and shallow aquatic biomes on our planet. Given these attributes, birds continue to be widely used as models for studies on population genetics, neurobiology, development and animal conservation. Moreover, domesticated birds contribute substantially to the World’s food source and economy as in the form of eggs, meat, ornament and culture. In terms of world health, the spread of avian influenza, West Niles virus and other zoonotic or emerging infectious diseases has led to the loss of human lives. As pests in agriculture, some species result in loss of billions of dollars per year. Clearly, ornithology has had and will continue to have a pronounced impact on both basic scientific research and on biosecurity of human society.
In the past decade, we have witnessed the rapid development of genome sequencing technologies and the resultant wealth of knowledge from animal genomes. The phylogeny of birds has been one of the most challenging vertebrate groups to decipher. Recently, our ordinal level project, a large international consortium led by researchers from BGI, University of Copenhagen, and Duke University, and including investigators from more than 20 countries, sequenced and/or collected the genomes from 48 bird species representing nearly all orders and covering a broad range of evolutionary diversity .The consortium was able to resolve much of the contested phylogenetic history of modern avian orders (Fig. 1). This project also addressed an extensive array of questions regarding avian evolution, diversity and behavior .Due to this success, we decided to expand our understanding and exploration of this unique class of vertebrates and extend these findings into multiple disciplines. To do so, the consortium has expanded and been reinforced through the addition of three more institutes as part of the leadership board: the Kunming Institute of Zoology and Institute of Zoology of Chinese Academy of Science in Beijing; the Smithsonian Institution in the USA; and the Center of Macroecology, Evolution and Climate in Denmark. They include experts in museum science, biogeography and ecology. Other contributing institutions and collaborators are listed in this website.
Given the small size and less complex features of bird genomes relative to other vertebrates, the ongoing advances in sequencing technologies, and the extensive availability of high quality tissue samples from birds deposited in museums around the world, reaching this ambitious goal is not only possible but also practical. In addition to contributing a large public genomic resource that will benefit the entire research community and human society, this project will include extensive collections of the morphological, physiological, ecological and behavioral traits of all species and will address a number of fundamental questions in evolution, ecology, and biodiversity.
The B10K project will allow the completion of a genomic level tree of life of the entire living avian class, decode the link between genetic variation and phenotypic variation, uncover the correlation of genetic evolutionary and biogeographical and biodiversity patterns across a wide-range of species, evaluate the impact of various ecological factors and human influence on species evolution, and unveil the demographic history of an entire class of organisms. Given all these aims, we are carrying out the project in three phases. Each phase focuses on the completion of milestones at hierarchical levels of avian classification (Fig. 2). We envision this project will have significant scientific and public impact that will change our understanding of avian biology and evolution, which in turn will affect our understanding of other organisms and open doors to new areas of research.

Figure 1. New reconstruction of the avian family tree, based on whole genome sequences of 48 bird species from 34 orders. See here for more. Painting by Jon Fjeldså.
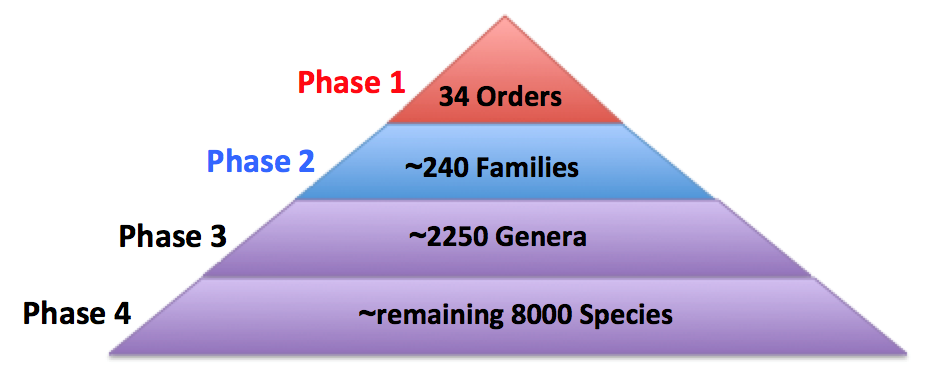
Figure 2. The B10K plan in four phases. Project phases are based on the hierarchical levels of avian classification. The ordinal level phase of c. 34 orders has been completed, and we are now generating genomic data for representatives of c. 240 families (plus several additional subfamilies). Sample collection is ongoing for Phase 3 (2250+ genera) and Phase 4 (the remaining 8000 species).
The milestones of B10K project
The B10K was built on the success of the Avian Phylogenomics Project, in which we carefully selected 48 bird genomes for genomic sequencing and comparative analyses. These bird species represent all 32 Neognathae and two of the five Palaeognathae orders. With these genomes, we performed the phylogenomics analyses to investigate the bird family, and applied comparative genomic methods to address various questions such as avian genome evolution, sex chromosome evolution, molecular basis of flight, loss of teeth, and vocal learning. In addition, to address the technical problems encountered in this project, we developed new bioinformatic methods and software tools. The findings of this project have been reported in ca. 50 research papers, including either papers in a Dec. 12th (2014) special issue of Science (Fig. 3).
On 12th November 2020, B10K Consortium announced the completion of its second milestone—the release of 363 genomes representing 92% of all bird families in Nature as a cover story (Fig. 4). We used this comparative genome dataset in combination with a pipeline that leverages a reference-free whole-genome alignment to identify orthologous regions in greater numbers than has previously been possible and to recognize genomic novelties in particular bird lineages. The densely sampled alignment provided a single-base-pair map of selection and revealed extensive patterns of weak selection in predominantly non-coding DNA. These results demonstrate that increasing the diversity of genomes used in comparative studies can reveal more shared and lineage-specific variation, and improve the investigation of genomic characteristics.
The latest breakthrough of the B10K project has published avian's family-level species tree. In this study, the affinities of birds are rearranged, Neornithae were divided into 4 groups, including the newly proposed group ‘elementaves’. New evidence from time estimate studies provides support for the ‘big bang’ scenario. Present genes telling ancient stories —— we have found a 21Mb region of neoavian genomes showing a consistent history for the first divergence among Neoaves circa 65 Mya.

Figure 3. Cover story in Science (Volume 346 Issue 6215, 12 December 2014). Image credit: Flip De Nooyer/Foto Natura/Minden Pictures/Corbis

Figure 4. Cover story in Nature (Volume 587 Issue 7833, 12 November 2020). Image credit: SciStories.